Exploring the PanNuke Dataset with PyTorch Implementations from Scratch

Nuclei segmentation in histopathology images is a foundational task in computational pathology. This project was created to be both a powerful tool and an open educational resource. It provides a complete ecosystem for training and evaluating deep learning models on the PanNuke dataset, which contains over 7,000 annotated image patches from 19 different tissue types.
Whether you're a student exploring computer vision and seeking PyTorch implementations, or a medical practitioner interested in interacting with this dataset, this repository offers a transparent and hands-on approach. Its core philosophy is to demystify these models by providing everything from the data preprocessing pipeline to an interactive web application for inference.
A key feature of this project is the collection of well-known segmentation architectures, all coded from the ground up in PyTorch to maximize understanding and customizability. The available models include:
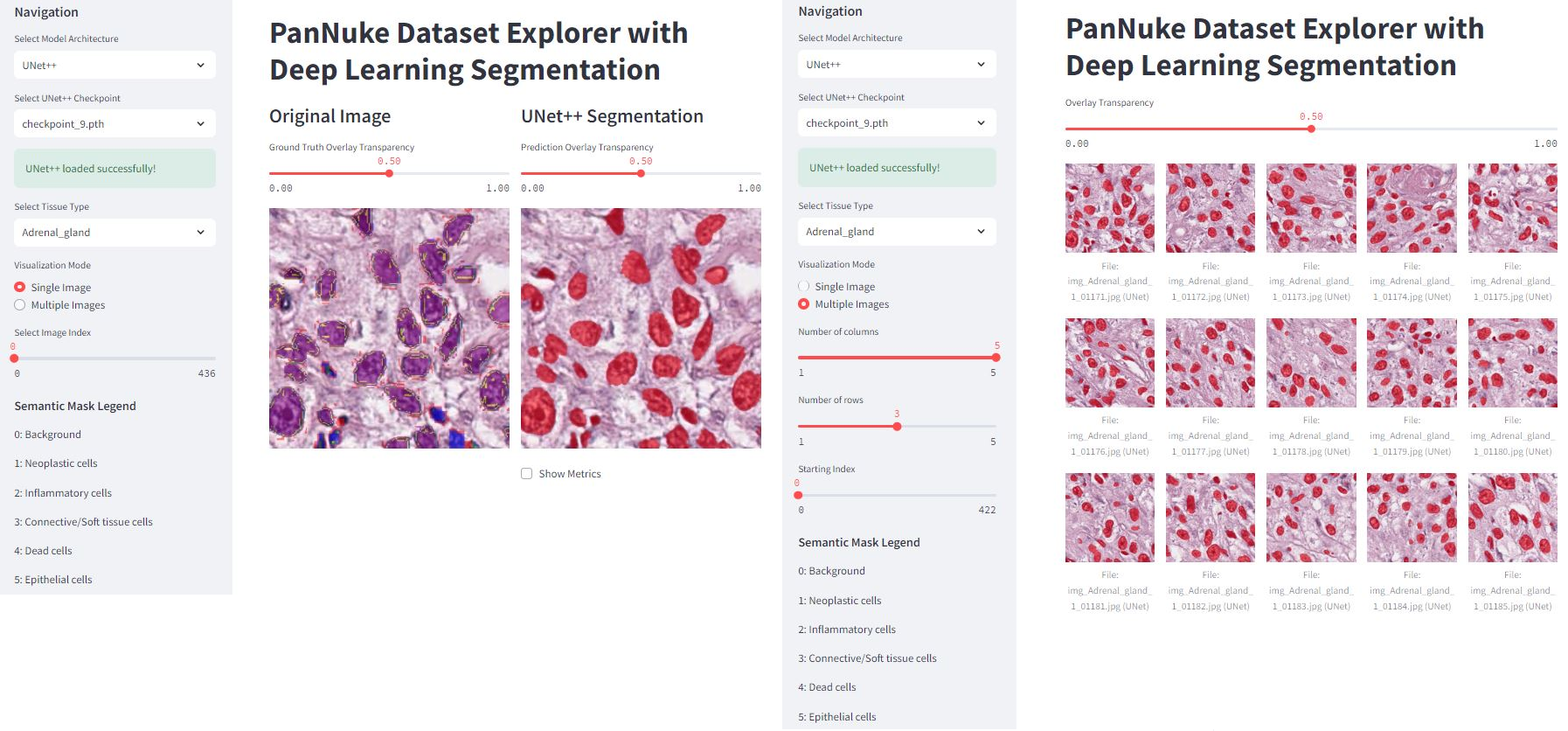
To make model exploration intuitive and accessible, the project includes a web application built with Streamlit. This interface allows you to:
This hands-on tool bridges the gap between training a model and seeing its tangible results, making it ideal for demonstrations, debugging, and educational purposes.
The complete source code, training pipeline, and web application are available on GitHub. Dive in to train your own models or explore the pre-built implementations!